Protein Modification, Folding, Secretion, and Degradation
#WakeUp
😈💩👎
mRNA Processing
Eukaryotic pre-mRNA receives a 5′ cap and a 3′ poly (A) tail before introns are removed and the mRNA is considered ready for translation.
LEARNING OBJECTIVES
Outline the steps of pre-mRNA processing
KEY TAKEAWAYS
Key Points
A 7-methylguanosine cap is added to the 5′ end of the pre-mRNA while elongation is still in progress. The 5′ cap protects the nascent mRNA from degradation and assists in ribosome binding during translation.
A poly (A) tail is added to the 3′ end of the pre-mRNA once elongation is complete. The poly (A) tail protects the mRNA from degradation, aids in the export of the mature mRNA to the cytoplasm, and is involved in binding proteins involved in initiating translation.
Introns are removed from the pre-mRNA before the mRNA is exported to the cytoplasm.
Key Terms
intron: a portion of a split gene that is included in pre-RNA transcripts but is removed during RNA processing and rapidly degraded
moiety: a specific segment of a molecule
spliceosome: a dynamic complex of RNA and protein subunits that removes introns from precursor mRNA
Pre-mRNA Processing
The eukaryotic pre-mRNA undergoes extensive processing before it is ready to be translated. The additional steps involved in eukaryotic mRNA maturation create a molecule with a much longer half-life than a prokaryotic mRNA. Eukaryotic mRNAs last for several hours, whereas the typical E. coli mRNA lasts no more than five seconds.
Pre-mRNAs are first coated in RNA-stabilizing proteins; these protect the pre-mRNA from degradation while it is processed and exported out of the nucleus. The three most important steps of pre-mRNA processing are the addition of stabilizing and signaling factors at the 5′ and 3′ ends of the molecule, and the removal of intervening sequences that do not specify the appropriate amino acids. In rare cases, the mRNA transcript can be “edited” after it is transcribed.
5′ Capping
While the pre-mRNA is still being synthesized, a 7-methylguanosine cap is added to the 5′ end of the growing transcript by a 5′-to-5′ phosphate linkage. This moiety protects the nascent mRNA from degradation. In addition, initiation factors involved in protein synthesis recognize the cap to help initiate translation by ribosomes.
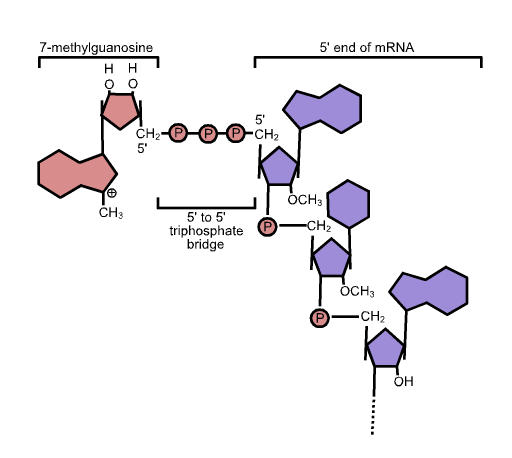
5′ cap structure: Capping of the pre-mRNA involves the addition of 7-methylguanosine (m7G) to the 5′ end. The cap protects the 5′ end of the primary RNA transcript from attack by ribonucleases and is recognized by eukaryotic initiation factors involved in assembling the ribosome on the mature mRNA prior to initiating translation.
3′ Poly-A Tail
While RNA Polymerase II is still transcribing downstream of the proper end of a gene, the pre-mRNA is cleaved by an endonuclease-containing protein complex between an AAUAAA consensus sequence and a GU-rich sequence. This releases the functional pre-mRNA from the rest of the transcript, which is still attached to the RNA Polymerase. An enzyme called poly (A) polymerase (PAP) is part of the same protein complex that cleaves the pre-mRNA and it immediately adds a string of approximately 200 A nucleotides, called the poly (A) tail, to the 3′ end of the just-cleaved pre-mRNA. The poly (A) tail protects the mRNA from degradation, aids in the export of the mature mRNA to the cytoplasm, and is involved in binding proteins involved in initiating translation.

Poly (A) Polymerase adds a 3′ poly (A) tail to the pre-mRNA.: The pre-mRNA is cleaved off the rest of the growing transcript before RNA Polymerase II has stopped transcribing. This cleavage is done by an endonuclease-containing protein complex that binds to an AAUAAA sequence upstream of the cleavage site and to a GU-rich sequence downstream of the cut site. Immediately after the cleavage, Poly (A) Polymerase (PAP), which is also part of the protein complex, catalyzes the addition of up to 200 A nucleotides to the 3′ end of the just-cleaved pre-mRNA.
Pre-mRNA Splicing
Eukaryotic genes are composed of exons, which correspond to protein-coding sequences (ex-on signifies that they are expressed), and intervening sequences called introns (int-ron denotes their intervening role), which may be involved in gene regulation, but are removed from the pre-mRNA during processing. Intron sequences in mRNA do not encode functional proteins.
Discovery of Introns
The discovery of introns came as a surprise to researchers in the 1970s who expected that pre-mRNAs would specify protein sequences without further processing, as they had observed in prokaryotes. The genes of higher eukaryotes very often contain one or more introns. While these regions may correspond to regulatory sequences, the biological significance of having many introns or having very long introns in a gene is unclear. It is possible that introns slow down gene expression because it takes longer to transcribe pre-mRNAs with lots of introns. Alternatively, introns may be nonfunctional sequence remnants left over from the fusion of ancient genes throughout evolution. This is supported by the fact that separate exons often encode separate protein subunits or domains. For the most part, the sequences of introns can be mutated without ultimately affecting the protein product.
Intron Processing
All introns in a pre-mRNA must be completely and precisely removed before protein synthesis. If the process errs by even a single nucleotide, the reading frame of the rejoined exons would shift, and the resulting protein would be dysfunctional. The process of removing introns and reconnecting exons is called splicing. Introns are removed and degraded while the pre-mRNA is still in the nucleus. Splicing occurs by a sequence-specific mechanism that ensures introns will be removed and exons rejoined with the accuracy and precision of a single nucleotide. The splicing of pre-mRNAs is conducted by complexes of proteins and RNA molecules called spliceosomes.
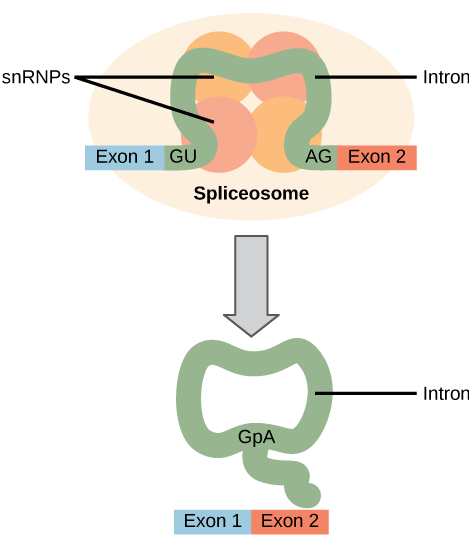
Pre-mRNA splicing: Pre-mRNA splicing involves the precise removal of introns from the primary RNA transcript. The splicing process is catalyzed by large complexes called spliceosomes. Each spliceosome is composed of five subunits called snRNPs. The spliceseome’s actions result in the splicing together of the two exons and the release of the intron in a lariat form.
Each spliceosome is composed of five subunits called snRNPs (for small nuclear ribonucleoparticles, and pronounced “snurps”.) Each snRNP is itself a complex of proteins and a special type of RNA found only in the nucleus called snRNAs (small nuclear RNAs). Spliceosomes recognize sequences at the 5′ end of the intron because introns always start with the nucleotides GU and they recognize sequences at the 3′ end of the intron because they always end with the nucleotides AG. The spliceosome cleaves the pre-mRNA’s sugar phosphate backbone at the G that starts the intron and then covalently attaches that G to an internal A nucleotide within the intron. Then the spliceosme connects the 3′ end of the first exon to the 5′ end of the following exon, cleaving the 3′ end of the intron in the process. This results in the splicing together of the two exons and the release of the intron in a lariat form.
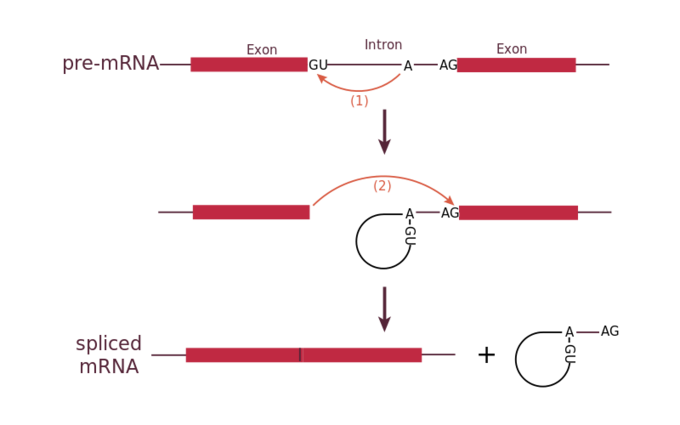
Mechanism of pre-mRNA splicing.: The snRNPs of the spliceosome were left out of this figure, but it shows the sites within the intron whose interactions are catalyzed by the spliceosome. Initially, the conserved G which starts an intron is cleaved from the 3′ end of the exon upstream to it and the G is covalently attached to an internal A within the intron. Then the 3′ end of the just-released exon is joined to the 5′ end of the next exon, cleaving the bond that attaches the 3′ end of the intron to its adjacent exon. This both joins the two exons and removes the intron in lariat form.
Denaturation and Protein Folding
Denaturation is a process in which proteins lose their shape and, therefore, their function because of changes in pH or temperature.
LEARNING OBJECTIVES
Discuss the process of protein denaturation
KEY TAKEAWAYS
Key Points
Proteins change their shape when exposed to different pH or temperatures.
The body strictly regulates pH and temperature to prevent proteins such as enzymes from denaturing.
Some proteins can refold after denaturation while others cannot.
Chaperone proteins help some proteins fold into the correct shape.
Key Terms
chaperonin: proteins that provide favorable conditions for the correct folding of other proteins, thus preventing aggregation
denaturation: the change of folding structure of a protein (and thus of physical properties) caused by heating, changes in pH, or exposure to certain chemicals
Each protein has its own unique sequence of amino acids and the interactions between these amino acids create a specify shape. This shape determines the protein’s function, from digesting protein in the stomach to carrying oxygen in the blood.
Changing the Shape of a Protein
If the protein is subject to changes in temperature, pH, or exposure to chemicals, the internal interactions between the protein’s amino acids can be altered, which in turn may alter the shape of the protein. Although the amino acid sequence (also known as the protein’s primary structure) does not change, the protein’s shape may change so much that it becomes dysfunctional, in which case the protein is considered denatured. Pepsin, the enzyme that breaks down protein in the stomach, only operates at a very low pH. At higher pHs pepsin’s conformation, the way its polypeptide chain is folded up in three dimensions, begins to change. The stomach maintains a very low pH to ensure that pepsin continues to digest protein and does not denature.
Enzymes
Because almost all biochemical reactions require enzymes, and because almost all enzymes only work optimally within relatively narrow temperature and pH ranges, many homeostatic mechanisms regulate appropriate temperatures and pH so that the enzymes can maintain the shape of their active site.
Reversing Denaturation
It is often possible to reverse denaturation because the primary structure of the polypeptide, the covalent bonds holding the amino acids in their correct sequence, is intact. Once the denaturing agent is removed, the original interactions between amino acids return the protein to its original conformation and it can resume its function.
However, denaturation can be irreversible in extreme situations, like frying an egg. The heat from a pan denatures the albumin protein in the liquid egg white and it becomes insoluble. The protein in meat also denatures and becomes firm when cooked.

Denaturing a protein is occasionally irreversible: (Top) The protein albumin in raw and cooked egg white. (Bottom) A paperclip analogy visualizes the process: when cross-linked, paperclips (‘amino acids’) no longer move freely; their structure is rearranged and ‘denatured’.
Chaperone proteins (or chaperonins ) are helper proteins that provide favorable conditions for protein folding to take place. The chaperonins clump around the forming protein and prevent other polypeptide chains from aggregating. Once the target protein folds, the chaperonins disassociate.
Protein Folding, Modification, and Targeting
In order to function, proteins must fold into the correct three-dimensional shape, and be targeted to the correct part of the cell.
LEARNING OBJECTIVES
Discuss how post-translational events affect the proper function of a protein
KEY TAKEAWAYS
Key Points
Protein folding is a process in which a linear chain of amino acids attains a defined three-dimensional structure, but there is a possibility of forming misfolded or denatured proteins, which are often inactive.
Proteins must also be located in the correct part of the cell in order to function correctly; therefore, a signal sequence is often attached to direct the protein to its proper location, which is removed after it attains its location.
Protein misfolding is the cause of numerous diseases, such as mad cow disease, Creutzfeldt-Jakob disease, and cystic fibrosis.
Key Terms
prion: a self-propagating misfolded conformer of a protein that is responsible for a number of diseases that affect the brain and other neural tissue
chaperone: a protein that assists the non-covalent folding/unfolding of other proteins
Protein Folding
After being translated from mRNA, all proteins start out on a ribosome as a linear sequence of amino acids. This linear sequence must “fold” during and after the synthesis so that the protein can acquire what is known as its native conformation. The native conformation of a protein is a stable three-dimensional structure that strongly determines a protein’s biological function. When a protein loses its biological function as a result of a loss of three-dimensional structure, we say that the protein has undergone denaturation. Proteins can be denatured not only by heat, but also by extremes of pH; these two conditions affect the weak interactions and the hydrogen bonds that are responsible for a protein’s three-dimensional structure. Even if a protein is properly specified by its corresponding mRNA, it could take on a completely dysfunctional shape if abnormal temperature or pH conditions prevent it from folding correctly. The denatured state of the protein does not equate with the unfolding of the protein and randomization of conformation. Actually, denatured proteins exist in a set of partially-folded states that are currently poorly understood. Many proteins fold spontaneously, but some proteins require helper molecules, called chaperones, to prevent them from aggregating during the complicated process of folding.
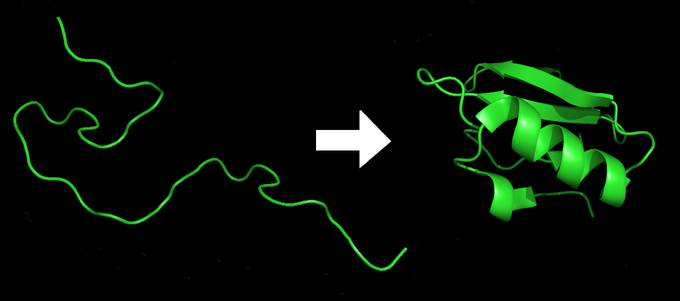
Protein folding: A protein starts as a linear sequence of amino acids, then folds into a 3-dimensional shape imbued with all the functional properties required inside the cell.
Protein Modification and Targeting
During and after translation, individual amino acids may be chemically modified and signal sequences may be appended to the protein. A signal sequence is a short tail of amino acids that directs a protein to a specific cellular compartment. These sequences at the amino end or the carboxyl end of the protein can be thought of as the protein’s “train ticket” to its ultimate destination. Other cellular factors recognize each signal sequence and help transport the protein from the cytoplasm to its correct compartment. For instance, a specific sequence at the amino terminus will direct a protein to the mitochondria or chloroplasts (in plants). Once the protein reaches its cellular destination, the signal sequence is usually clipped off.
Misfolding
It is very important for proteins to achieve their native conformation since failure to do so may lead to serious problems in the accomplishment of its biological function. Defects in protein folding may be the molecular cause of a range of human genetic disorders. For example, cystic fibrosis is caused by defects in a membrane-bound protein called cystic fibrosis transmembrane conductance regulator (CFTR). This protein serves as a channel for chloride ions. The most common cystic fibrosis-causing mutation is the deletion of a Phe residue at position 508 in CFTR, which causes improper folding of the protein. Many of the disease-related mutations in collagen also cause defective folding.
A misfolded protein, known as prion, appears to be the agent of a number of rare degenerative brain diseases in mammals, like the mad cow disease. Related diseases include kuru and Creutzfeldt-Jakob. The diseases are sometimes referred to as spongiform encephalopathies, so named because the brain becomes riddled with holes. Prion, the misfolded protein, is a normal constituent of brain tissue in all mammals, but its function is not yet known. Prions cannot reproduce independently and not considered living microoganisms. A complete understanding of prion diseases awaits new information about how prion protein affects brain function, as well as more detailed structural information about the protein. Therefore, improved understanding of protein folding may lead to new therapies for cystic fibrosis, Creutzfeldt-Jakob, and many other diseases.
Regulating Protein Activity and Longevity
A cell can rapidly change the levels of proteins in response to the environment by adding specific chemical groups to alter gene regulation.
LEARNING OBJECTIVES
Explain how chemical modifications affect protein activity and longevity
KEY TAKEAWAYS
Key Points
Proteins can be chemically modified by adding methyl, phosphate, acetyl, and ubiquitin groups.
Protein longevity can be affected by altering stages of gene regulation, including but not limited to altering: accessibility to chromosomal DNA for transcription, rate of translation, nuclear shuttling, RNA stability, and post-translational modifications.
Ubiquitin is added to a protein to mark it for degradation by the proteasome.
Key Terms
ubiquitin: a small polypeptide present in the cells of all eukaryotes; it plays a part in modifying and degrading proteins
proteasome: a complex protein, found in bacterial, archaeal and eukaryotic cells, that breaks down other proteins via proteolysis
Chemical Modifications, Protein Activity, and Longevity
Proteins can be chemically modified with the addition of methyl, phosphate, acetyl, and ubiquitin groups. The addition or removal of these groups from proteins regulates their activity or the length of time they exist in the cell. Sometimes these modifications can regulate where a protein is found in the cell; for example, in the nucleus, the cytoplasm, or attached to the plasma membrane.
Chemical modifications occur in response to external stimuli such as stress, the lack of nutrients, heat, or ultraviolet light exposure. These changes can alter protein function, epigenetic accessibility, transcription, mRNA stability, or translation; all resulting in changes in expression of various genes. This is an efficient way for the cell to rapidly change the abundance levels of specific proteins in response to the environment. Because proteins are involved in every stage of gene regulation, the phosphorylation of a protein (depending on the protein that is modified) can alter accessibility to the chromosome, can alter translation (by altering transcription factor binding or function), can change nuclear shuttling (by influencing modifications to the nuclear pore complex), can alter RNA stability (by binding or not binding to the RNA to regulate its stability), can modify translation (increase or decrease), or can change post-translational modifications (add or remove phosphates or other chemical modifications). All of these protein activities are affected by the phosphorylation process. The enzymes which are responsible for phosphorylation are known as protein kinases. The addition of a phosphate group to a protein can result in either activation or deactivation; it is protein dependent.
Another example of chemical modifications affecting protein activity include the addition or removal of methyl groups. Methyl groups are added to proteins via the process of methylation; this is the most common form of post-translational modification. The addition of methyl groups to a protein can result in protein-protein interactions that allows for transcriptional regulation, response to stress, protein repair, nuclear transport, and even differentiation processes. Methylation on side chain nitrogens is considered largely irreversible while methylation of the carboxyl groups is potentially reversible. Methylation in the proteins negates the negative charge on it and increases the hydrophobicity of the protein. Methylation on carboxylate side chains covers up a negative charge and adds hydrophobicity. The addition of this chemical group changes the property of the protein and, thus, affects it activity.
The addition of an ubiquitin group to a protein marks that protein for degradation. Ubiquitin acts like a flag indicating that the protein lifespan is complete. These proteins are moved to the proteasome, an organelle that functions to remove proteins to be degraded. One way to control gene expression is to alter the longevity of the protein: ubiquitination shortens a protein’s lifespan.
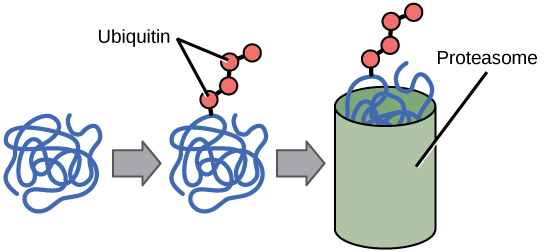
Ubiquitin Tags: Proteins with ubiquitin tags are marked for degradation within the proteasome.
LICENSES AND ATTRIBUTIONS


